Drug repositioning, or putting already approved compounds to new uses, has gained ground in recent years as a cost-saving alternative to traditional drug development. Before directly testing on animals or humans to see if an old drug will do new tricks, researchers begin with computational methods to generate hypotheses and narrow the number of repositioning candidates. But how are these computational methods checked for accuracy?
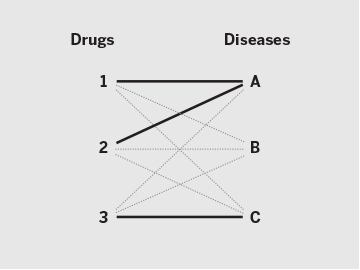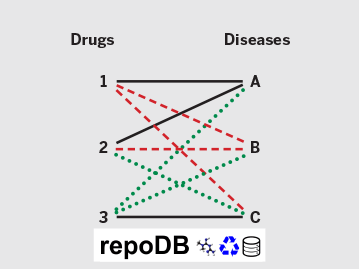
Suppose three drug–disease combinations have succeeded so far (solid lines): Drugs 1 and 2 are approved for Disease A, and Drug 3 for Disease C. For analytical validation purposes, many computational drug repositioning methods assume that the remaining six combinations (gray dotted lines) are failures, or “false positives.” With repoDB, investigators can instead access actual drug failure data, or “true negatives” (red dashed lines). This provides not only a more reliable benchmark for one’s methods but also concrete insight into drug discovery avenues yet to be explored (green dotted lines).
In fact, a systematic review of this question had not been undertaken until recently, when PhD candidate Adam Brown, working on his thesis in the lab of Assistant Professor Chirag Patel, took on the challenge. As they reported in the October 2016 Briefings in Bioinformatics, Brown and Patel found wide variation in validation methods.
They also identified a key weakness of the most popular of these (sensitivity- and specificity-based validation, or SSV): specificity should be a measure of truly negative results found in actual drug-indication pairs, but this data has not been readily available for consideration. In its absence, investigators have instead assumed all unknowns (i.e., the unannotated drug–indication pairs) to be false positives for purposes of calculating specificity. This meant the same drug-indication pairs that were being identified as holding therapeutic potential by a given computational repositioning method were, perhaps ironically, also being considered false when validating the very same method.
A solution Brown and Patel proposed was the creation of a “gold standard” database of both true positives and true negatives. That database is repoDB, which they released this month and describe in Nature's Scientific Data.
In repoDB, failed drug-indication pairs are drawn from ClinicalTrials.gov by way of the American Association of Clinical Trials database. Approved drugs are drawn from the DrugCentral database. Both failed and approved drugs link directly to their DrugBank records, and failed drugs also link to their corresponding trial records, so researchers can quickly drill deeper for specifics. The database can be searched and filtered as well as downloaded in part or as a whole.
With this new gold standard database, investigators have not only a more reliable benchmark for their computational methods but also extra insight into drug discovery avenues that have yet to be explored.