HuBMAP (the Human BioMolecular Atlas Program) has released its inaugural data for use by the scientific community and the general public. Included in this release are detailed, 3D anatomical data and genetic sequences of healthy tissues from seven organ types, at the level of individual cells as well as many bulk tissue datasets. HuBMAP’s ultimate goal is to provide the framework required for scientists to create a 3D atlas of the human body.
HuBMAP’s tools and maps are openly available for research to accelerate understanding of the relationships between cell and tissue organization and function, as well as human health. Visitors can find the data at portal.hubmapconsortium.org.
HuBMAP is a consortium of 18 diverse collaborative research teams across the United States and Europe. HuBMAP values secure, open sharing and collaboration with other consortia and the wider research community. The consortium is managed by a trans-National Institutes of Health (NIH) working group of staff from the NIH Common Fund; National Heart, Lung, and Blood Institute; National Institute of Biomedical Imaging and Bioengineering; and the National Institute of Diabetes and Digestive and Kidney Diseases.
How HuBMAP Will Help People
A human adult is made up of trillions of cells, and how those cells interact, connect, and arrange into tissues has a direct effect on our health. HuBMAP will create the next generation of molecular data acquisition technologies and computational tools, enabling the generation of foundational 3D tissue maps and construction of an atlas of the function and relationships among cells in the human body. These maps and atlas can lead us to a better understanding of how the relationships among our cells affects our health.
Real-world applications in which potential HuBMAP data users have expressed interest include:
- A surgeon would like to use HuBMAP to visualize tissues to explain disease processes and surgical interventions to patients.
- A software developer would like to use HuBMAP data to build software tools for doctors to create novel solutions to help people with specific medical conditions.
- A computational biologist would like to use HuBMAP to understand how differences between individual cells in the body lead to normal development—or progression of disease.
- A drug developer would like to use HuBMAP to identify where in the body a particular gene is highly expressed, to find new treatment uses for a drug candidate in development.
- A patient diagnosed with BRCA1-linked breast cancer would like to use HuBMAP to understand how variables such as age, sex and diagnosis interact.
- A parent would like to use HuBMAP to show their kids “cool biology work so that I can amaze, entertain, and educate” them.
First Data Release
The data in the first HuBMAP release include:
- De-identified donor, tissue sample, and assay data and metadata for the heart, kidney, large intestine, lymph nodes, small intestine, spleen, and thymus.
- Over 300 datasets contributed by HuBMAP research teams at CalTech, Stanford, University of California, San Diego, University of Florida and Vanderbilt.
- Assays include microscopy, mass spectrometry, and sequencing data with several assay types available for each modality.
- For additional information, see the list of assays details as well as donor, tissue sample, and assay metadata.
With the inaugural data release, HuBMAP will engage other atlas-creating consortia to instrumentally advance biomedical data sharing. Functions include:
- Spatial common coordinate framework data for select kidney and spleen datasets
- Faceted search for select metadata fields
- A cell-naming future-functionality preview
- Receiving feedback from the user community for subsequent releases
- Protocols, Github repositories and APIs linked from the portal
Future data releases will likely include data from, among other assays, Cell DIVE, Micro/Nano CT, MIBI, DART-FISH, Light Sheet, and Immuno-SABER.
The Gehlenborg Lab and the Kharchenko Lab in the Department of Biomedical Informatics at Harvard Medical School are funded by the National Institutes of Health Common Fund, grants OT2OD026677 and U54HL145608. Future releases of data will follow, beginning in late 2020 or early 2021. For more information about these releases, data access, or the data themselves, see hubmapconsortium.org or email info@hubmapconsortium.org.
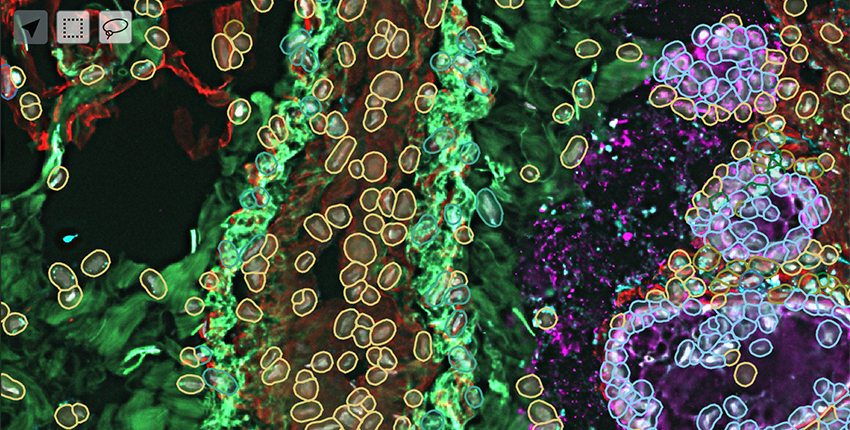
Sample images from the HuBMAP Portal's data visualization platform